Background: the expansion of high-dimensional cytometry, single-cell, and imaging technologies has lead to a significant shift in our approach to analysing immunological data. As the size and complexity of high-dimensional cytometry data continue to expand, comprehensive, scalable, and methodical computational analysis approaches are essential. Yet, contemporary clustering and dimensionality reduction tools alone are insufficient to analyze or reproduce analyses across large numbers of samples, batches, or experiments. Moreover, approaches that allow for the integration of data across batches or experiments are not well incorporated into computational toolkits to allow for streamlined workflows, nor are approaches to analyse time-series data.
Our team: to address these challenges, we formed the Immune Dynamics team, a multi-disciplinary collaboration between the Sydney Cytometry Facility and multiple research and technology labs at the University of Sydney, integrating expertise in immunology, infectious disease, high-dimensional single-cell & imaging technologies, computational biology, and data science.
Our work: collectively, our work involves the development of various high-dimensional cytometry, single-cell, and imaging methods, as well as advanced computational analysis approaches, to address challenges in modern single-cell and imaging sciences. With our collaborators, we apply these approaches to comprehensively map dynamic immune responses at the single-cell level over time and across multiple biological tissues, seeking to reveal key mediators of inflammation and immunity. In particular, we apply these approaches to the study of immunology and infectious disease, including emerging pathogens such as SARS-CoV-2 and COVID-19, Zika virus encephalitis, and West Nile virus encephalitis. We then explore how these datasets, technologies, and analysis methdologies may contribute to, or benefit from, efforts such as the Human Cell Atlas (HCA).
Below we describe areas of research. More information on the research interests of our individual members can be found on the team page.
Computational analysis and data integration across cytometry and single-cell technologies
A major focus of our collaborative group is the development of computational analysis solutions for high-dimensional cytometry data.
- Important – ability to test and compare different clustering, DR, and alignment/integration tools – in Spectre, make it easy to run multiple tools to optimiset he approach
- Inspire by efforts of HCA
- Sought to create an interface that would allow a seamless transition across a wide variety of single-cell data types – including flow, spectral, CyTOF, and scRNAseq, including multiomic assays such as CITE-seq and Abseq
- In particular faciliate integration and label transfer between datasets, so that reference datasets generated by the HCA and other groups could be leveraged in a wide variety of contexts, in both HD cytometry and single-cell SEQ
- –> by creating a platform and data-agnostic platform, we can efficiently incorporate various analysis tools, and particular integration tools, developed for cytometry (e.g. FlowSOM) and transcriptomics (e.g. Harmony, Liger, Seurat)
- By doing this, multiple approaches can be efficiently run and compared
–> in our case, focusing on methods that can scale to large datasets consisting of millions of cells (ref https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1850-9#Abs1)
Development
- Spectre for computational analysis
- Batch alignment/integration
- Paretobench and DR dev
- ICB paper example
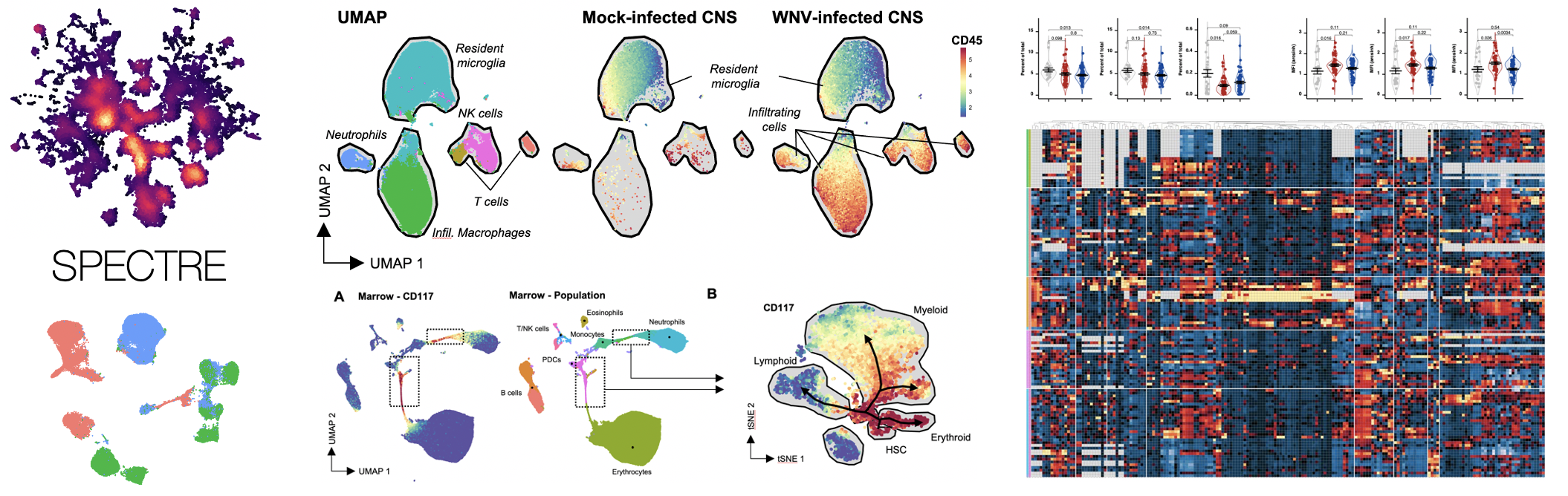
Application — examples in WNV/ZIKV/COVID etc
Time-series clustering analysis for mapping dynamic immune responses
- Another major focus is the incorporation of time-series analysis approaches, as this is an under-developed field, but critical in many areas on biology
Development
- ChronoClust
- TrackSOM
Application — examples in COVID etc
Spatial analysis of high-dimensional imaging data
- Spatial analysis platform –> integrated with Spectre
- Improved cell segmentation workflows
- Spatial statistics and neighbourhood analysis — examples in TBC
Community contributions and activitiers
- Roadshow examples
- Workshops
- HCA…
TBC: Development of high-dimensional cytometry and spatial technologies

Critical to investigating complex immune responses to disease is the capacity to measure cells at the single-cell level, examining multiple features of the cell simultaneously. To address this, we develop and apply a variety of high-dimensional cytometry technologies and assays. We have developed novel technology platforms, such as our world first 10-laser ‘LSR-X’ platform developed with BD, and one of the first published 25 colour flow cytometry assays (Ashhurst et al. 2017). We also developed the first assays to be used on Australia’s first mass cytometer (CyTOF) (Ashhurst et al. 2019), as well as Australia’s first Imaging Mass Cytometer (IMC, SpectreMAP Github). We have recently published a book (HM McGuire, TM Ashhurst (eds). 2019), featuring a collecting of up-to-date and cutting-edge protocols in mass cytometry, which is now a standard resource in the field. Most recently, we have implemented high-dimensional spectral cytometry platforms and panels, and performed comprehensive evaluations of both conventional and spectral cytometry (Niewold, Ashhurst et al. 2020).
ADD SCRNASEQ.
This research work is summarised in the following pages:
- High-dimensional cytometry and imaging technologies
- Computational and spatial analysis approaches
- Application to disease (incl. COVID-19)
- Engagement in the Human Cell Atlas (HCA) and other communities
An overview of this work can be found in this Oz Single Cell webinar 2020 on computational biology and this Fluidigm webinar 2020, featured on the ‘COVID-19 resources’ page.