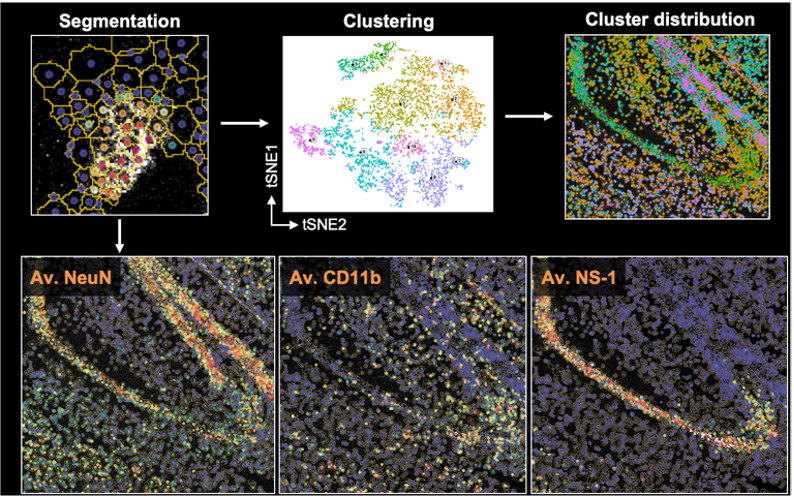
Spectre enables the analysis of high-dimensional imaging data, including data generated using Fluidigm’s Hyperion Imaging Mass Cytometer (IMC). Our current workflows support a basic (using CellProfiler) or comprehensive (using Ilastik) cell and region segmentation approach, followed by cellular and spatial analysis using FlowJo or Spectre (in R). For those gettings started with analysis, first check out our getting started page for installation instructions and some basic tutorials.
Please note: the original functions and workflows from SpectreMAP have now been directly incorporated in Spectre.
Overview
|
Introduction to cell segmentation and spatial analysis Here we provide background information on methods of analysing IMC data. GO TO PAGE |
Image visualisation and TIFF export

|
Image visualisation and TIFF export Here we provide instructions for initial image visualisation and TIFF export using MCD Viewer or HistoCat++. From here, TIFF files can be exported for use in our segmentation approaches. GO TO PAGE |
Cell segmentation
Here we provide protocols for performing initial cell segmentation.
|
Boundary-based multicut segmentation with Ilastik
|
Simple nuclear expansion segmentation with CellProfiler
|
Modified Bodenmiller lab segmentation approach
|
|
|

|

|
| Some IMC images contain extremely dense collections of cells, where cytoplasmic (and sometimes) nuclear signal from one cell is difficult to distinguish from another. In this protocol we describe boundary-based segmentation using the 'multi-cut' workflow in Ilastik. Additional cell type and region masks can be included, which dramatically enhances analysis. | This is the simplest form of cellular segmentation and analysis. The nuclear signal is identified, and the boundary is expanded outwards by a certain number of pixels. This then becomes the boundary of the cell mask. Although a very simplistic approach, only nuclear signal is required. | In this approach, the user trains a classifier to identify and predict 'nuclear', 'cytoplasmic', or 'background' pixels. This can be used to create more comprehensive cell masks than can be achieved using simple pixel expansion methods. This is based on the Bodenmiller lab workflow described here. Additional cell type and region masks can be included, which dramatically enhances analysis. |
Spatial analysis
|
Advanced spatial analysis workflow using Spectre (following Ilastik multicut segmentation)
|
Simple spatial analysis workflow using Spectre (following CellProfiler nuclear expansion segmentation)
|
Spatial data analysis using FlowJo
|


|


|


|
| An analysis workflow in R using Spectre that facilitates simultaneous cellular and spatial analysis following boundary-based segmentation with the multicut module of Ilastik.. | An analysis workflow in R using Spectre that facilitates simultaneous cellular and spatial analysis following nuclear-expansion segmentation in CellProfiler. | A workflow for analysing IMC data using FlowJo, after initial conversion of TIFF files and masks into FCS files. |
Other software and protocols
|
HistoCat (GUI, Matlab)
|
CytoMapper (R)
|
CytoMAP (GUI, Matlab)
|
|

|

|

|
| HistoCat software from the Bodenmiller/Shapiro labs. | CytoMapper software from the Bodenmiller lab. | CytoMAP software from the Gerner lab. |