2020-10-28 - 'Integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre' published
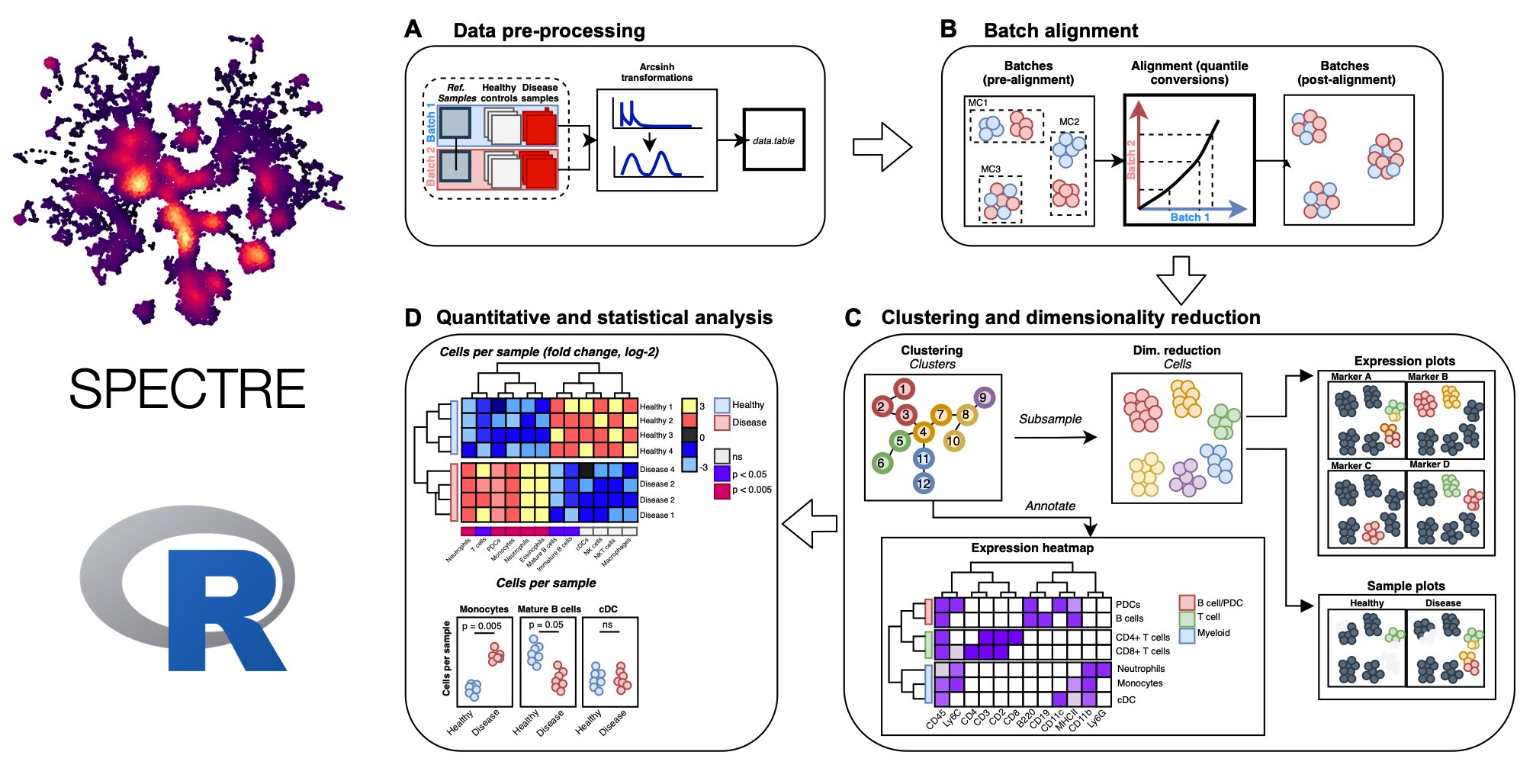
Spectre is an R package that enables comprehensive end-to-end integration and analysis of high-dimensional cytometry data from different batches or experiments. Spectre streamlines the analytical stages of raw data pre-processing, batch alignment, data integration, clustering, dimensionality reduction, visualisation and population labelling, as well as quantitative and statistical analysis. To manage large cytometry datasets, Spectre was built on the data.table framework – this simple table-like structure allows for fast and easy processing of large datasets in R. Critically, the design of Spectre allows for a simple, clear, and modular design of analysis workflows, that can be utilised by data and laboratory scientists. For more information, check out our ‘about Spectre’ page.
Code/download: Github page.
Manuscript: bioRxiv.
Presentation: Oz Single Cell webinar series, 2020.
Thrilled to announce our preprint is out on bioRxiv: https://t.co/YebSZu94Yq. Here we present Spectre, an R package that enables comprehensive end-to-end integration and analysis of high-dimensional cytometry data from different batches or experiments. https://t.co/l1crJst3lO https://t.co/hcs2rZd8Lk pic.twitter.com/IX4sa9K9Jf
— Dr Thomas Ashhurst (@TomAsh_1) October 26, 2020