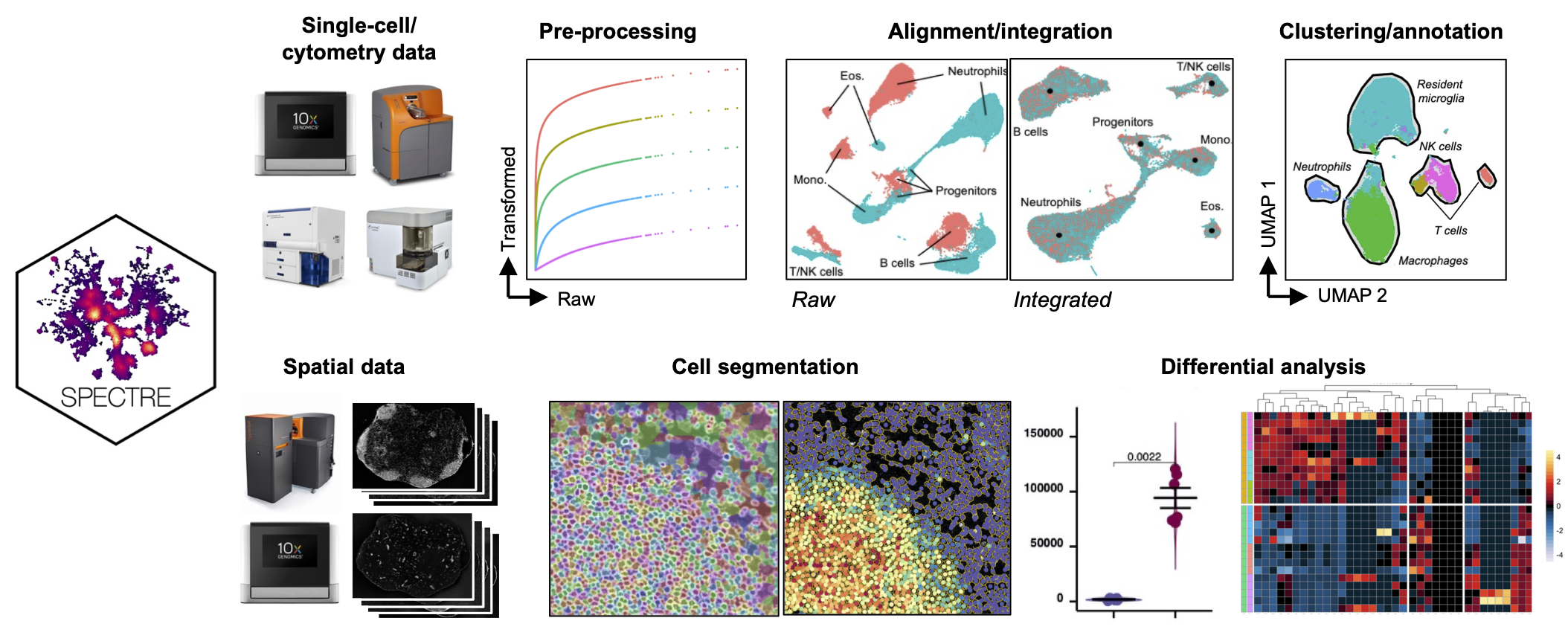
Spectre is an R package and computational toolkit that enables comprehensive end-to-end integration, exploration, and analysis of high-dimensional cytometry, spatial/imaging, or single-cell data from different batches or experiments. Spectre streamlines the analytical stages of raw data pre-processing, batch alignment/integration, clustering, dimensionality reduction, visualisation, population annotation, and quantitative/statistical analysis; with a simple, clear, and modular design of analysis workflows, that can be utilised by both data and laboratory scientists. For more information see our about page and check our our metrics page for how Spectre has been used in the community. To get started, check out our getting started, protocols, or tutorial pages below.
News
See our latest version
To receive updates, you can join our mailing list here.
- 2024-07-19: v1.2.0 release major updates to many functions and introduction of CI.
- 2023-11-12: v1.1.0 release minor updates to remove outdated dependencies.
- 2021-09-03: v1.0.0 release including major updates to our spatial functions and workflows.
- 2021-06-04: Spatial v0.5.0 release - analysis functions and workflows from SpectreMAP now incorporated directly in Spectre.
- 2021-04-10: Spectre manuscript published online in Cytometry A
- 2021-03-30: Updated list of papers that have referenced Spectre (and/or CAPX and associated scripts).
- 2021-03-08: Spectre v0.4.0 release – including significant improvements, as well as the addition of new batch alignment functions.
- 2020-10-23: Spectre pre-print now online: Ashhurst TM, Marsh-Wakefield F, Putri GH, et al. (2020). bioRxiv.
- 2019-07-31: Spectre Github repository established.
- 2019-05-11: The ‘CAPX’ workflow (the initial version of Spectre) published as part of a methods chapter of ‘Mass Cytometry: Methods and Protocols’ book: Ashhurst TM et al. (2019).
- 2018-09-25: CAPX Github established.
Getting started

|
Getting started with R, RStudio, and Spectre Here we provide instructions for installing R, RStudio, and Spectre, along with introductory tutorials for getting started. GO TO PAGE
|
Protocols and tutorials
|
Cytometry (flow, mass, spectral)
|
Spatial and imaging
|
Single-cell genomics
|

|

|

|
| Here we provide protocols and instructions for using Spectre for the integration and analysis of high-dimensional flow, spectral, or mass (CyTOF) cytometry data. | Here we provide protocols and instructions for using Spectre for the spatial analysis of Imaging Mass Cytometry (IMC) data. | Here we provide analysis options and tools to support scRNAseq analysis, in conjunction with existing tools such as Seurat and SingleCellExperiment. |