Overview
Spectre is an R package and computational toolkit that enables comprehensive end-to-end integration, exploration, and analysis of high-dimensional cytometry, spatial/imaging, or single-cell data from different batches or experiments. Spectre streamlines the analytical stages of raw data pre-processing, batch alignment/integration, clustering, dimensionality reduction, visualisation, population annotation, and quantitative/statistical analysis; with a simple, clear, and modular design of analysis workflows, that can be utilised by both data and laboratory scientists. The Spectre package was developed by Thomas Ashhurst (the Sydney Cytometry Core Research Facility, The University of Sydney and Centenary Institute), Felix Marsh-Wakefield (the School of Medical Sciences, The University of Sydney), and Givanna Putri (the School of IT, The University of Sydney); with support from Alanna Spiteri, Dr. Diana shinko, Dr. Mark Read, Dr. Adrian Smith, and Prof. Nicholas King.
Key publications:
- v0: Ashhurst et al, 2019, Mass Cytometry: Methods and Protocols (referred to at the time as ‘CAPX’)
- v1: Ashhurst et al, 2021, Cytometry A
Metrics
Our software and associated analysis protocols have been cited in at least 91 publications since 2018, both with our team and independently, in prestigious journals such as Nature, Cell, Nature Immunology, and Blood. It is also a featured analysis package for the Human Cell Atlas project. Spectre has been used in the study of COVID-19 pathogenesis, vaccine responses, NK cell biology, multiple myeloma, pediatric burns, multiple sclerosis, bone marrow transplantation, and cerebral malaria, among others.
See our usage statistics and a full list of publications citing Spectre here
Background
As the size and complexity of high-dimensional cytometry data continues to expand, comprehensive analysis tools that can scale to large datasets are required. In our early mass cytometry work, the analysis of high-dimensional datasets using manual methods became prohibitive, requiring the use of computational analysis approaches, such as clustering and dimensionality reduction. However, the analysis tools available at the time were not well suited to the analysis of large cytometry datasets, consistings of tens to hundreds of millions of single cells. Furthermore, popular clustering and dimensionality reduction tools alone are insufficient for scalable or reproducible integration of data across batches, experiments, or different cytometry/single-cell technologies. To address these limitations, we initially developed the ‘Cytometry Analysis Pipeline for large and compleX datasets’ (CAPX, the initial version of Spectre), an analysis workflow using the R programming language (Ashhurst et al. 2019), and derivative stand-alone scripts (e.g. ‘tSNEplots’). Following a seed funding grant from the Marie Bashir Institute for Infectious Diseases and Biosecurity, we established the ‘Immune Dynamics’ team, a collaborative multi-disciplinary group with a focus on the development of novel computational analysis tools to address challenges in high-dimensional analysis, with a particular focus on the analysis of large datasets, and the incorporation of data integration strategies. We solidified the functionality of the CAPX workflow into a package and re-named it ‘Spectre’. This was initially released as a pre-print in bioRxiv (Ashhurst*, Marsh-Wakefield*, Putri* et al. bioRxiv. 2020) and then published in Cytometry A (Ashhurst*, Marsh-Wakefield*, Putri* et al. Cytometry A. 2020).
What is Spectre
Spectre is an R package designed to facilitate data analysis workflows that simplify and streamline data manipulation and annotation, population identification (clustering, classification), and dimensionality reduction (t-SNE, UMAP) etc in high-dimensional cytometry data. Strategic implementation of batch-alignment, data-integration, and cell-type classification tools allow for the integrated analysis of multiple experiments, as well as a reproducible system for rapid and repeated cell type identification in large datasets. Critically, the fundamental data structures used within Spectre, along with the implementation of classifiers allow for the scalable analysis of very large high-dimensional datasets. In addition to high-dimensional cytometry datasets, we’ve also developed functions to allow for spatial analysis of high-dimensional imaging datasets, such as those generated by Imaging Mass Cytometry. The simple, clear, and modular design of analysis workflows allow for these tools to be used by informaticians and laboratory scientists alike.
Along with the various R packages used within Spectre, some key packages such as the cytofkit and Seurat R packages provided inspiration for elements of the package design. Our team develops other computational approaches, including tools for the analysis of time-series data (e.g. ChronoClust, Putri et al 2019; and TrackSOM, Koutsakos et al 2020) or spatial data (e.g. SpectreMAP). As our tools reach a stable level of development, we incorporate these into Spectre, so they may form part of a cohesive workflow.
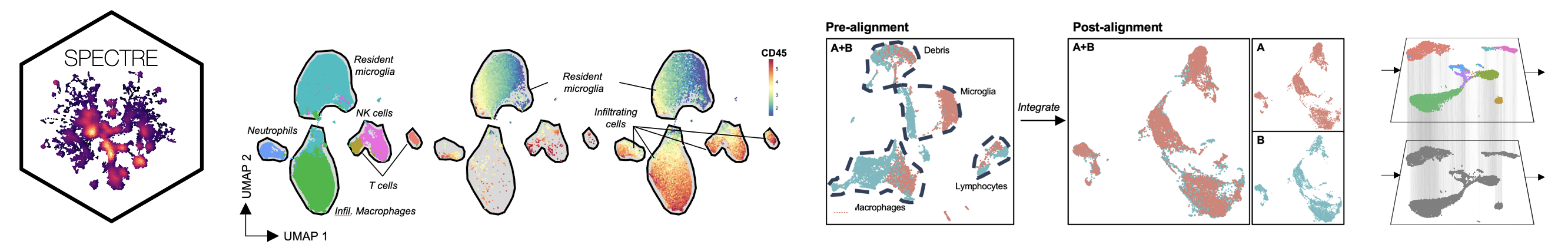
Along with flow, spectral, or mass cytometry data, Spectre enables spatial analysis of Imaging Mass Cytometry (IMC) data. Through our extension package, ‘SpectreMAP’, we can import, manage, and visualise TIFF files using RStudio. Once cell segmentation has been performed (using our protocols, or those developed by others), the marker expression data for each cell across multiple images can be calculated and incorporated into a single data.table.
In the following presentation, we describe the integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre in detail, as part of the Oz Single Cell seminar series 2020.
Spectre built on data.table
Many existing computational tools store data in a custom format, such as the flowFrame or SingleCellExperiment object, that provide excellent field-specific structuring of single-cell data. In Spectre, data management and operations are performed using the data.table format, an extension of R’s base data.frame, provided by the data.table package. This table-like structure organises cells (rows) against cellular features or metadata (columns). This simple data.table structure allows for the high-speed processing, manipulation (subsetting, filtering, etc.), and plotting of large datasets, as well as fast reading/writing of large CSV files.
Rather than storing analysis outputs (clusters, dimensionality reduction values, annotations etc) in separate areas of a custom data format, Spectre simply adds new columns to the existing data.table. The simplicity of this data structure facilitates extremely fast and simple filtering/subsetting by data.table, as every cell (row) contains all of the information relevant for that cell: such as cellular expression, samples/groups, clusters/populations, and dimensionality reduction coordinates.
Clustering and dimensionality reduction strategies for large datasets
Whilst clustering tools such as FlowSOM scale well to large datasets, dimensionality reduction approaches such as t-SNE and UMAP do not; as they incur lengthy computing time, excessive memory usage, and significant crowding effects that inhibit their utility. Whilst some improvements to runtime (flt-SNE) and plot crowding (opt-SNE) have been made, scalability and plot crowding limitations persist. As dimensionality reduction tools are primarily used to visualise cellular data and clustering results, we plot a subset of the clustered data, which addresses scalability and retains legibility. By using proportional subsampling from each sample, the relative number of cells from each cluster in each sample can be preserved in a smaller dataset, allowing for interpretable analysis via DR. Putative cellular populations amongst the clusters can then be identified, and annotated in both the subsampled DR dataset, as well as the whole clustered dataset. The whole annotated dataset can subsequently be used in downstream quantification and statistical analysis.